Curious how CancerHallmarks.com works?
In this brief video, we guide you through the key features of the platform, demonstrate how to input your gene list, explain how to interpret hallmark enrichment results, and show you how to download publication-ready plots. Whether you're a researcher or student, this quick guide will help you make the most of our tool.
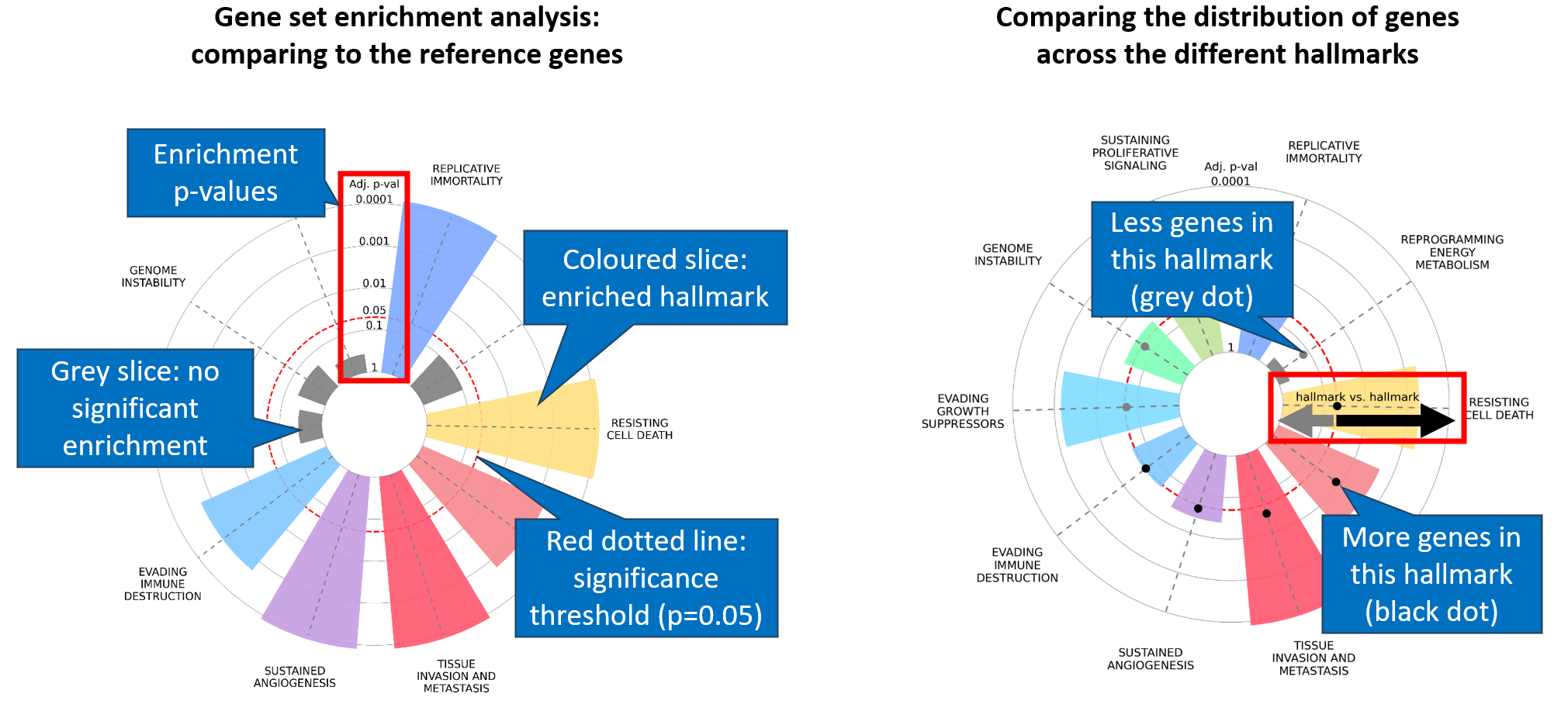
What does the hallmark enrichment plot show?
The first layer of data represents the enrichment of the cancer hallmarks when compared to the reference set of genes. In this overrepresentation analysis, differently colored slices represent each of the ten cancer hallmarks, and only the significant ones (adjusted p < 0.05) are colored. The size of the slices corresponds to the strength of the enrichment compared to the chosen reference cancer hallmark gene set.
A second layer of information compares the distribution of genes across the different hallmarks to each other. The value of the Hallmark vs Hallmark equals the actual number of genes linked to the selected hallmark in the investigated gene set divided by the expected number of genes in this selected hallmark. The expected number of genes is calculated by dividing the total genes in the selected hallmark by the sum of all genes linked to each hallmark and multiplied by the sum of all genes linked to all hallmarks in the investigated gene set. Black or grey dots provide a graphical representation for each of the ten hallmarks. The black dot is proportionally closer to the external rim if one hallmark has comparatively more genes. Hallmarks with no genes have a grey dot at the inner line.
What is the difference between the integrated and the core set?
We created a database of candidate cancer hallmark genes by extracting and merging gene sets from seven resources. The resulting complete list is termed “Integrated cancer hallmark gene set” and consists of 6,763 individual genes altogether. Most genes were referenced in only one of the seven resources, while 1,574 genes were included in at least two gene lists. This shorter list is termed the „Core cancer hallmark gene set”.
What are the advantages of CancerHallmarks compared to Gene Ontology?
- Obvious link to a biological function
- Cancer-specific genes
- Facilitates data integration and comparison
- Graphical visualization of the outcome
 Uncover overrepresented cancer hallmarks
Uncover overrepresented cancer hallmarks